RNA Polymerase
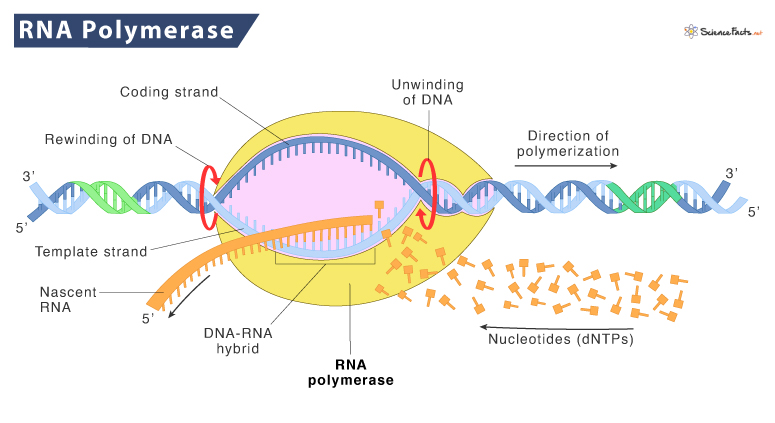
RNA polymerase (RNA Pol or RNAP) is the enzyme that synthesizes RNA from a template of DNA, a process known as transcription. It is thus a DNA-dependent RNA polymerase. The messenger RNAs (mRNAs) produced from transcription are further used to encode functional proteins or make non-coding RNAs, such as tRNA, rRNA, and miRNA.
RNAPs not only transcribe DNA but are also responsible for proofreading the newly synthesized RNA strand and taking part in the recognition of the transcription start and termination sites.
American biochemist Roger D. Kornberg was awarded the Nobel Prize in chemistry in 2006 for demonstrating the images of the enzyme during the various stages of transcription.
Types and Structure of RNA Polymerases
RNAP is a multisubunit enzyme found in prokaryotes, eukaryotes, and viruses. While bacteria and archaea contain only one RNAP, eukaryotes have many RNAPs: RNAP I, RNAP II, RNAP III, RNAP IV, and RNAP Vs.
Despite having different types, the basic structure and its mode of operation are conserved throughout all living organisms. They require zinc and magnesium as cofactors.
Prokaryotic RNA Polymerase
In bacteria, a single RNA polymerase transcribes all types of RNAs. The core enzyme (ββ′α2ω) consists of five subunits: two alpha (α) subunits of 36 kDa, a beta (β) subunit of 150 kDa, a beta prime subunit (β′) of 155 kDa, and a small omega (ω) subunit. When the sigma (σ) factor binds to the core, it forms the holoenzyme. The five subunits are described below:
- β′ is the largest subunit that is encoded by the rpoC gene. It is part of the active center responsible for RNA synthesis. Also, it contains some elements for non-sequence-specific interactions with DNA and nascent RNA.
- β is the second large subunit and is encoded by the rpoB gene. It contains the other part of the active center responsible for RNA synthesis and the rest of the elements for non-sequence-specific interactions with DNA and nascent RNA.
- α is the third largest subunit, having two copies: α’ and α”. Each α subunit has an N-terminal αNTD domain and a C-terminal αCTD domain. αNTD helps assemble the enzyme, and αCTD recognizes and interacts with the promoter and the upstream DNA elements. α subunit also contains elements for interaction with the regulatory factors.
- ω is the smallest of all subunits that helps assemble and provide stability to the core enzyme.
To bind to the promoter, the core RNAP binds to the sigma (σ), forming the holoenzyme (ββ′α2ω σ) with 6 subunits. The σ factor reduces the core enzyme’s affinity for binding to non-specific DNA sequences while binding to the promoter. It allows transcription to initiate at the correct site.
Like bacteria, archaea have a single type of RNAP. Structurally, it is similar to bacterial RNAP and eukaryotic RNAP II.
Eukaryotic RNA Polymerase
Eukaryotes have many different nuclear RNAPs, and each recognizes a different promoter sequence, synthesizing some form of the RNA or the other.
- RNA polymerase I (RNAP I) synthesizes pre-rRNA 45S, which matures into the ribosome’s 28S, 18S, and 5.8S subunits.
- RNA polymerase II (RNAP II) produces precursors of mRNAs and most snRNA and microRNAs.
- RNA polymerase III (RNAP III) synthesizes tRNAs, rRNA 5S, and other small RNAs found in the nucleus and cytosol
- RNA polymerase IV (RNAP IV) produces siRNA in plants.
- RNA polymerase V (RNAP V) has RNAs involved in siRNA-directed heterochromatin formation in plants.
Chloroplast in eukaryotic cells contains an RNAP similar to bacterial RNAP. The sigma (σ) factor of the RNA polymerase is encoded by nuclear genes.
How RNA Polymerase Works
RNAP is an abundant enzyme that catalyzes the phosphodiester bonds linking nucleotides by hydrolyzing pyrophosphate from nucleoside triphosphates to form a linear mRNA chain. The growing RNA chain is extended one nucleotide at a time in the 5’→3’ direction using nucleoside triphosphates (ATP, CTP, UTP, and GTP).
Unlike DNA polymerase, RNAP has increased infidelity. On average, one error is made every 10,000 nucleotides. RNAP has a proofreading mechanism to find and replace the wrongly incorporated ribonucleotide and replacing them with the correct one.
What is the Function of RNA Polymerase
Here are the primary purposes of RNA polymerase in a cell:
Perform Transcription
The RNA polymerase is primarily responsible for transcription, the process by which a template DNA strand is used to form RNA. RNA polymerase produces all types of RNA, including messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), microRNA (miRNA), and even non-coding RNAs and ribozymes.
Once RNA polymerase attaches to the promoter (where RNA polymerase begins transcribing a gene), it moves along the DNA template in the 5 to 3’ direction, adding nucleotides to the growing RNA chain. This elongation process is similar to building a chain of RNA letters based on the instructions from the DNA template.
Through the regulation of transcription, cells control gene expression. Adapting to the changing environment, maintaining basic metabolic processes, and performing intracellular functions depend on gene expressions.
Promoter Recognition
Before transcription can begin, the initial target of the RNA polymerase is to locate the specific region of the DNA called the promoter. Promoters are start sites for transcription. Sigma factors in RNA polymerase help them to recognize these promoters.
Terminate Transcription
After transcribing the gene, RNA polymerase sends a termination signal to the DNA. This signal tells the enzyme to stop transcription, releasing the newly formed RNA molecule.
RNA Polymerase vs. DNA Polymerase
RNA polymerase differs from DNA polymerase in many ways:
| Basis | RNA Polymerase | DNA Polymerase |
|---|---|---|
| 1. Function | Perform transcription of DNA (making RNA copies of the template DNA) | Perform replication of DNA (making copies of the parent DNA) |
| 2. Product | RNA | DNA |
| 3. Cell Cycle Phase | Required in G1 and G2-phases of interphase | Required in S-phase of interphase |
| 4. Primer Requirement In Initiation | Need a primer for initiation of transcription | Does not need a primer for initiation of replication |
| 5. Base Pairs Used | Adenine, Guanine, Cytosine, and Uracil | Adenine, Guanine, Cytosine, and Thymine |
| 6. Error Rate | Very high | Very low |
| 7. Proofreading Activity | Absent | Present |
-
References
Article was last reviewed on Tuesday, November 7, 2023