Polymerase Chain Reaction
Polymerase chain reaction, known as PCR, is an experimental technique used to produce millions and millions of copies of DNA or RNA (nucleic acid) samples. It was developed by Kary Mullis and his colleagues in the 1980s, around the time the Human Genome Project was being planned.
Principle
It is based on the process of DNA replication, where a specific DNA polymerase known as taq polymerase, isolated from the bacterium Thermus aquaticus, is utilized. This enzyme creates a new DNA strand that is complementary to the template strand by incorporating nucleotides onto the existing 3′-OH group of the growing DNA strand.
Because DNA polymerase can only attach nucleotides to the end of a strand, it requires a short segment of DNA, called a primer, to which it can attach the incoming nucleotide. As a result, it enables us to amplify a specific segment of DNA within a limited timeframe.
Components
To perform PCR, we need the following:
- The DNA template contains the specific sequence that is being amplified.
- DNA polymerase is the enzyme that makes the complementary strand during DNA replication. Taq DNA polymerase is mostly used for its ability to remain stable at high temperatures.
- Oligonucleotide primer is a short segment of single-stranded DNA (approximately 20 nucleotides long) that is complementary to the template strand.
- Deoxyribonucleotide triphosphates (dNTPs) provide the energy to run this process.
- The buffer system, composed of potassium and magnesium ions, provides the environment to carry out the process.
All these components are mixed in a tube and introduced into a PCR machine, which provides the ideal conditions for amplification to occur.
How Does It Work
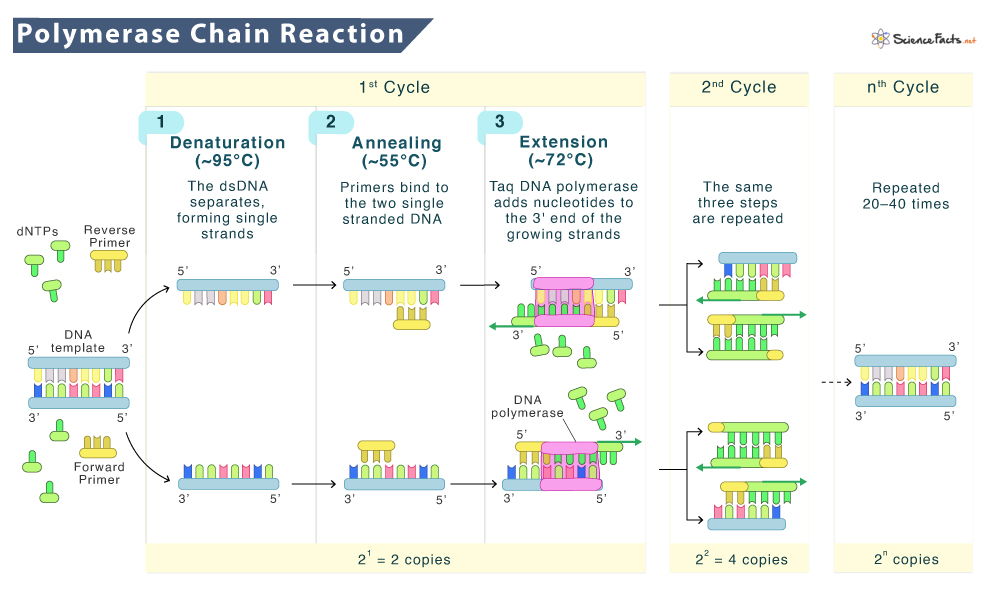
The three basic steps of PCR are:
- Denaturation: The double-stranded DNA is heated to almost 95°C to split the hydrogen bonds between the base pairs so that the DNA strand is opened up (turned into a single-strand).
- Annealing: The temperature is lowered to almost 55°C, which helps the primers to bind to their corresponding, complementary single strands. It is the step that actually decides the region of amplification. There are two primers (forward and reverse each) for two single strands.
- Extension: The temperature is then raised to 72°C, and the incoming nucleotides add themselves to the 3’ end of the primer by Taq polymerase. This elongation occurs in the 5’ to 3’ direction, with the polymerase adding almost 1000 base pairs per minute.
These three steps are repeated 20 to 30 times to generate the amplified product. The entire process takes 2 to 4 hours in total, depending on the size of the DNA that is being copied. The amplification process works through exponential growth, where the DNA made in each cycle is used as a template to make the DNA in the next cycle, thereby increasing the DNA content twofold in each step.
Types
PCR is not a one-size-fits-all technique. Its different types serve specific purposes.
- Quantitative (qPCR): Also known as real-time PCR, it measures the quantity of DNA amplified in real time using fluorescent probes.
- Reverse Transcription (RT-PCR): It converts RNA into complementary DNA (cDNA) using the enzyme reverse transcriptase before amplification.
- Hot-Start: Modifies the PCR process by using a heat-activated DNA polymerase, thus reducing non-specific amplification and improving the output.
- Nested: Involves two rounds of PCR to increase specificity. The first round amplifies a broad target, while the second round is more specific and acts on a region within the first product.
- Multiplex: Amplifies multiple targets in a single PCR reaction by using multiple sets of primers.
- In Situ PCR: Combines PCR with in situ hybridization, allowing the amplification of DNA directly within tissues or cells on a slide.
Applications
PCR provides the necessary copies of DNA templates for various applications.
- Medicine: Detecting genetic diseases, identifying pathogens like viruses and bacteria, and finding mutations
- Forensic: DNA fingerprinting in identifying criminals and in paternity testing
- Research: Cloning DNA for sequencing, studying gene expression, and finding genetic variations in populations
Limitations
Although it is a widely used technique in molecular biology, it has certain limitations:
- PCR amplifies DNA exponentially at each step. However, due to different factors like resource limitations, accumulation of pyrophosphate molecules, self-annealing of the accumulating product, and the presence of some inhibitors in the sample, the PCR reaction reaches a ‘plateau effect,’ which makes the quantification unreliable. However, in such a situation, we can use real-time quantitative PCR.
- The specificity of PCR depends on primers. Poorly designed primers can result in non-specific amplification or primer-dimer formation, which reduces the efficiency and accuracy of the reaction.
- PCR is generally effective for amplifying DNA fragments that are 3 to 5 kb in length. While amplifying longer sequences, the efficiency decreases.
-
References
Article was last reviewed on Wednesday, September 25, 2024