Operon
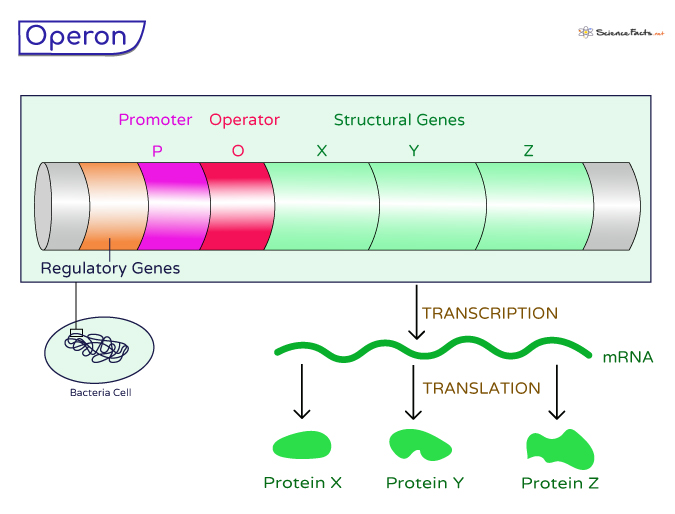
An operon is the functional unit of genetic regulation found in prokaryotic cells, such as bacteria. It consists of a cluster of genes that work together as a single unit to give a single messenger RNA (mRNA) molecule, which then encodes multiple proteins. The two most widely studied operons are the lactose (lac) operon and tryptophan (trp) operon in Escherichia coli (E. coli).
François Jacob and Jaques Monod first proposed the operon model of gene regulation in the lac operon of E. coli. Since then, scientists have discovered the parts that control the functions of an operon. While operons occur primarily in prokarytes, they are rare in eukaryotes but do exist.
Structure and Parts of an Operon
In prokaryotes, operons have several key components that work together to control gene expression efficiently. These components include:
1. Promoter (P)
The promoter is a crucial region of DNA located upstream of the structural genes within the operon. It serves as the initiation site for transcription, providing a binding site for RNA polymerase, the enzyme responsible for synthesizing RNA from the DNA template. RNA polymerase recognizes the promoter and attaches to it, initiating the transcription of the structural genes.
2. Operator (O)
The operator is another critical DNA sequence within the operon, positioned between the promoter and the structural genes. The operator acts as a regulatory switch that can turn the expression of the operon on or off. It controls access to the structural genes by interacting with specific regulatory proteins.
3. Structural Genes (X, Y, Z)
The structural genes are the actual genes within the operon that encode proteins essential for a particular biological function or pathway. These genes are organized in tandem and transcribed as a single mRNA molecule. Each structural gene codes for a specific protein, and their collective action is required for the complete function of the operon-regulated pathway.
4. Regulatory Genes
In addition to the components directly involved in transcription and translation, operons also involve regulatory genes located elsewhere in the bacterial genome. These regulatory genes code for regulatory proteins that influence the activity of the operon. There are two primary types of regulatory proteins: repressor and activator proteins.
a. Repressor Proteins are involved in negative regulation, inhibiting gene expression. When a repressor protein binds to the operator, it physically blocks RNA polymerase from accessing the promoter, preventing transcription of the structural genes. Inducible operons and repressible operons both use repressor proteins to control gene expression.
b. Activator Proteins are involved in positive regulation, enhancing gene expression. When an activator protein binds to a specific DNA sequence (enhancer), it facilitates the binding of RNA polymerase to the promoter, leading to increased transcription of the structural genes.
5. Effector Molecules
It includes inducers and corepressors.
Inducers are small molecules interacting with repressor proteins, causing a conformational change that renders the repressor inactive. It allows RNA polymerase to bind to the promoter, initiating transcription. Inducible operons rely on the presence of inducers to activate gene expression.
Corepressors are small molecules interacting with repressor proteins, promoting their binding to the operator. It blocks RNA polymerase from initiating transcription, effectively inhibiting gene expression. Repressible operons require the presence of corepressors to turn off gene expression when the pathway’s end product is in excess.
Inducible vs. Repressible Operon Regulation
There are two types of operon regulation:
In Inducible Operons
In inducible operons, the default state is ‘off.’ When a particular molecule, called an inducer, is present in the cell’s environment, it binds to the repressor protein. This binding renders the repressor inactive, allowing RNA polymerase to attach to the promoter to transcribe the genes. An example of an inducible operon is the lac operon, which controls lactose metabolism in E. coli.
In Repressible Operons
Repressible operons function oppositely. They are in an “on” state by default, and the genes are continuously transcribed. However, when a specific molecule, called a corepressor, is present, it binds to the repressor protein, activating it. The repressor then attaches to the operator, preventing RNA polymerase from initiating transcription. An example of a repressible operon is the trp operon, which is responsible for producing tryptophan biosynthetic enzymes in bacteria.
Functions of an Operon
Thus operons play a vital role in efficiently regulating gene expression based on the immediate needs of the cell, enabling prokaryotic organisms to adapt to their changing environment effectively.
-
References
Article was last reviewed on Tuesday, August 29, 2023